Tutorial 01: Adding a new monatomic species entry to your local data store.#
If you would like to do calculations using a plasma species that you haven’t used before, you will need to generate a minplascalc data entry for it first. You only need to do this once: minplascalc can store the species data in a plain-text file formatted using JSON syntax, and it will then be available for use in any of your future calculations.
Data needed to create a species can be obtained from the energy levels and lines section of the NIST Atomic Spectra Database, which can be found at http://physics.nist.gov/PhysRefData/ASD. For example, the landing page for levels looks like this:
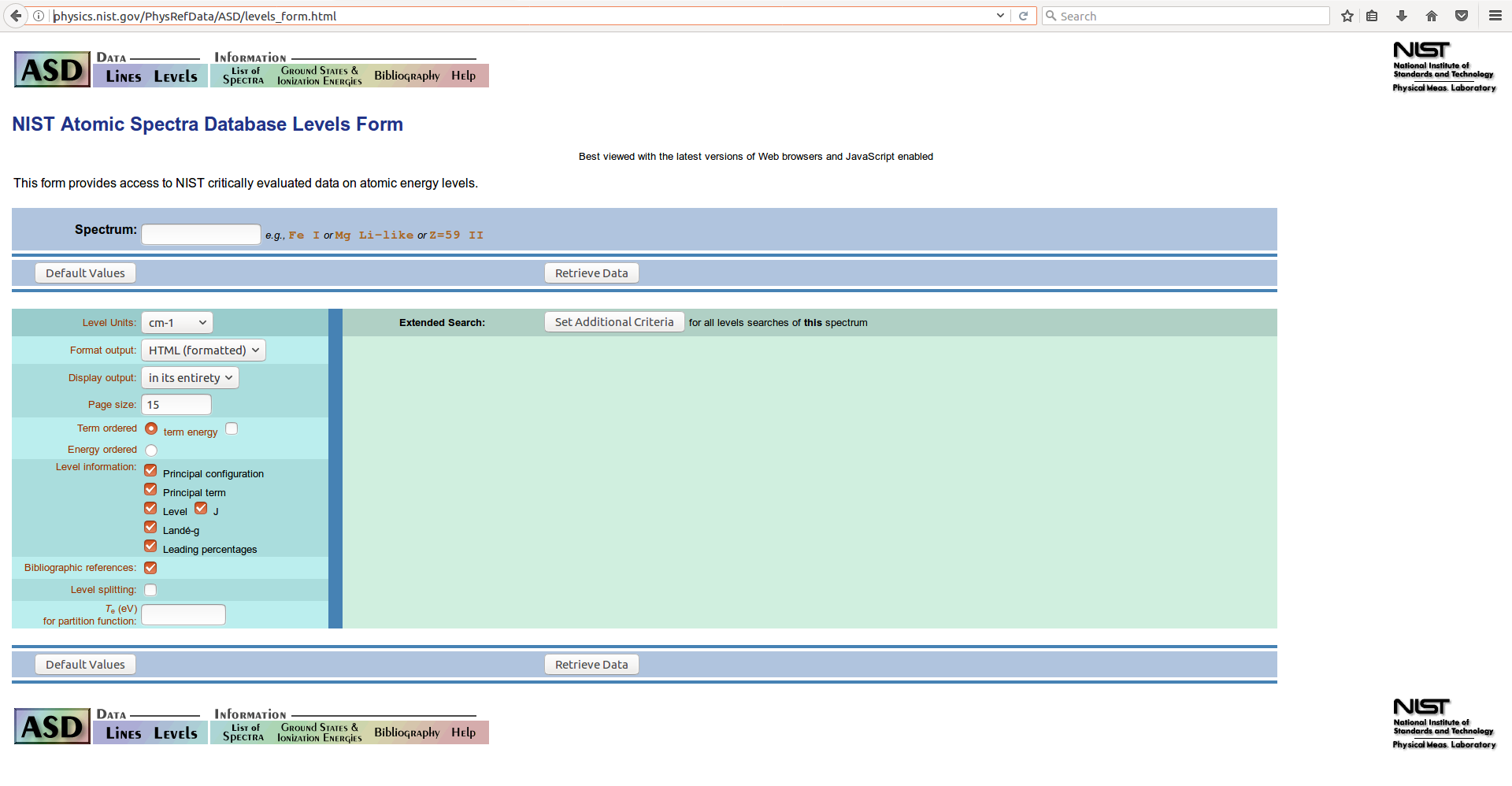
You must then specify the atom or ion that you want to retrieve energy level information for. Let’s get the data for the singly-charged oxygen cation species \(O^+\), which is “O II” in spectrographic terminology. Enter the identifier in the Spectrum field on the form:
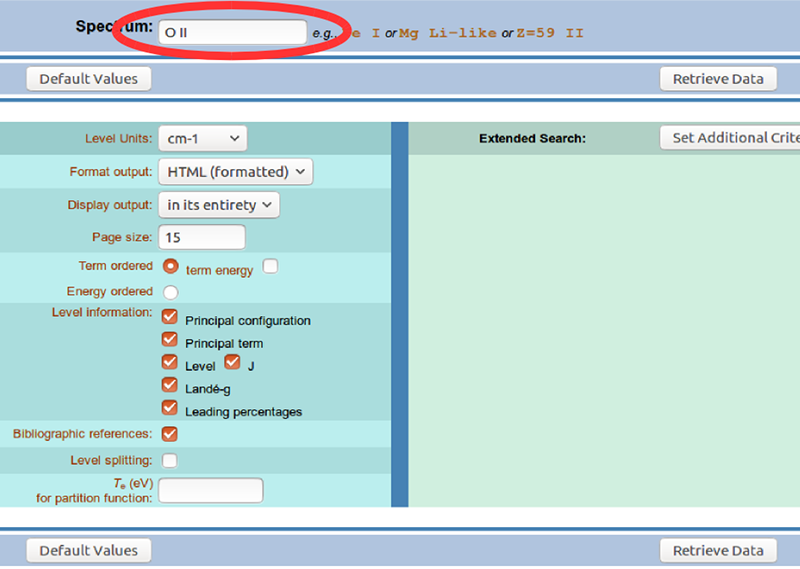
Set your preferences according to how you prefer to access the data. Make sure the “Level” and “J” checkboxes are ticked, since these are the data we will need for minplascalc.
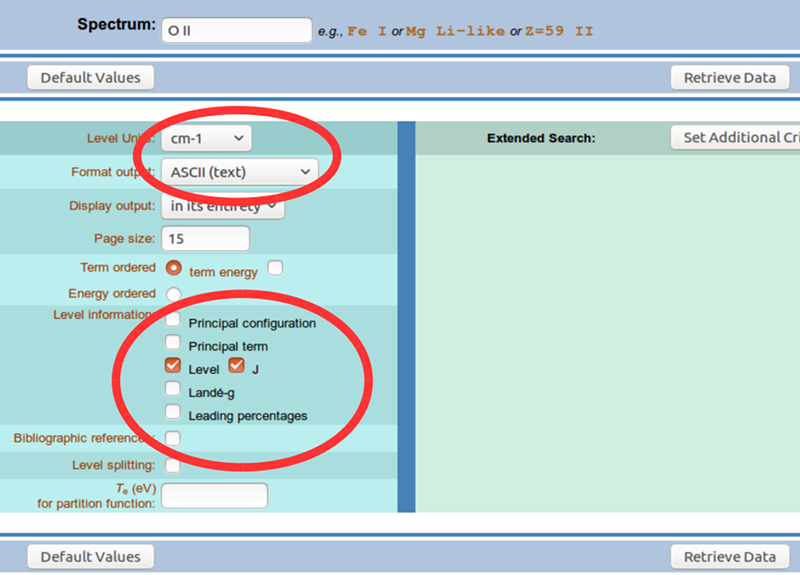
Click the Retrieve Data button. You should receive a page with the energy levels listed out. You’ll need to transfer this data into some format that you can manipulate in Python, either by saving text manually or using web scraping tools.


Convert the data to a list of length-2 lists, one for each eletronic energy level. Each list entry must contain the degeneracy of the level \(j\) and the level’s energy in J. A similar exercise must then be performed for the spectral lines using the “lines” form on the NIST ASD - these must be stored in a list of length-3 lists containing the line wavelength \(\lambda\) in m, the transition strength \(g_k A_{ki}\) in 1/s, and the line energy \(E_k\) in J. This exercise has already been performed for the oxygen cation, and data lists from NIST ASD are stored in pickle files at data/demo/nist.
Additional pieces of data needed to create a species object are:
The elemental stoichiometry of the species.
The molar mass of the species, in kg/mol.
The electric charge in units of the elementary charge.
The ionisation energy of the species, in J.
The polarisability of the species, in \(m^3\).
The multiplicity of the ground state.
The number of effective valence electrons, only used by neutral species (see docstring for more information).
Information about electron collision cross sections, only used by neutral species (see docstring for more information).
A list of sources describing the provenance of the data.
Running the following code snippet will create a minplascalc Species object for the \(O^+\) ion, and then store it to a file for later reuse:
import pickle
from minplascalc.species import Monatomic
from minplascalc.utils import get_path_to_data
with open(get_path_to_data("demo/nist/nist_Oplus_levels"), "rb") as f:
elevels = pickle.load(f)
with open(get_path_to_data("demo/nist/nist_Oplus_emission_lines"), "rb") as f:
elines = pickle.load(f)
oxygenplus = Monatomic(
name="O+",
stoichiometry={"O": 1},
molar_mass=0.01599885642,
charge_number=1,
ionisation_energy=5.6270249236e-18,
energy_levels=elevels,
polarisability=0.391e-30,
multiplicity=4,
effective_electrons=None,
electron_cross_section=None,
emission_lines=elines,
sources=[
"NIST Atomic Spectra Database (ver. 5.3), [Online]. A Kramida, "
"Yu Ralchenko, J Reader, and NIST ASD Team, National Institute "
"of Standards and Technology, Gaithersburg MD., "
"http://physics.nist.gov/asd"
],
)
print(oxygenplus)
oxygenplus.to_file()
Species: O+
Type: Monatomic ion
Stoichiometry: {'O': 1}
Molar mass: 0.01599885642 kg/mol
Charge number: 1
Ionisation energy: 5.6270249236e-18 J
Energy levels: 275
Polarisability: 3.91e-31 m^3
Multiplicity: 4
Effective valence electrons: None
Electron cross section data: None
Emission lines: 857
What’s happening here? First we import the minplascalc package, then unpack the NIST data files for energy levels and emission lines, then we create a Monatomic species object, and finally we save the contents to a data file
The Monatomic class constructor takes as arguments the lists of energy levels and lines, as well as the pieces of data described above. Once created, the object can be written out to disk using the Species object’s to_file utility function, which saves data to a human-readable JSON-formatted file. It takes an optional file path argument - if omitted, the filename is created using the species’ name and written to the current working directory.
After this process it will be possible to create an \(O^+\) species object in any minplascalc calculation by importing it using either the explicit path to the JSON file, or (preferably) just the name of the species provided the JSON file is stored in any of the standard minplascalc data paths - see later tutorials for examples. Note that for this to work correctly in all cases, minplascalc’s JSON files must be stored on a case-sensitive operating system or partition.
Total running time of the script: (0 minutes 0.015 seconds)