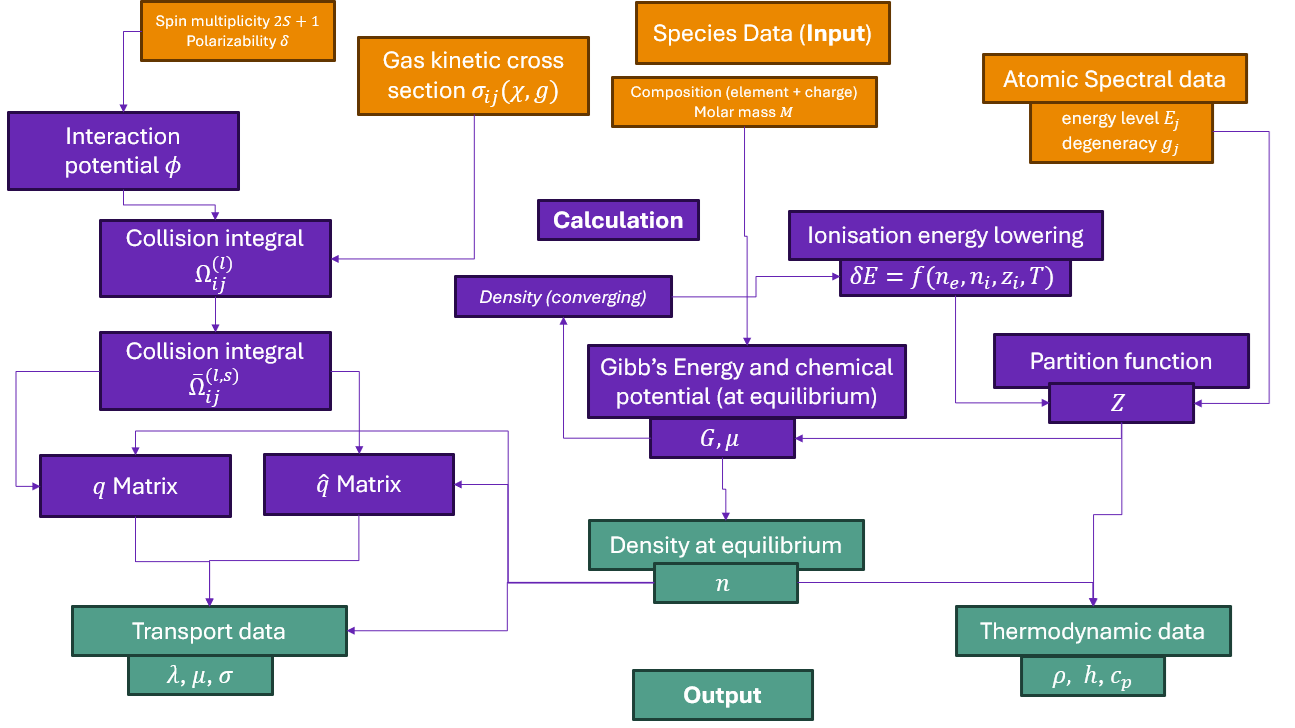
Tutorial 02: Adding a new diatomic species entry to your local data store.#
If you would like to do calculations using a plasma species that you haven’t used before, you will need to generate a minplascalc data entry for it first. You only need to do this once< - minplascalc can store the species data in a plain-text file formatted using JSON syntax, and it will then be available for use in any of your future calculations.
The procedure for generating a minplascalc data entry for a new diatomic species is entirely manual since the data required must be retrieved from various locations. The majority of data for a wide range of species is available from the NIST Chemistry WebBook, http://webbook.nist.gov/chemistry/form-ser/. Enter the formula for the diatomic species you’re interested in in the search box at the top of the page, and click the Search button. If you’re looking for data for charged diatomic ions, make sure the “Exclude ions from the search” box is unchecked.
Let’s build a data entry for Silicon Monoxide, SiO:
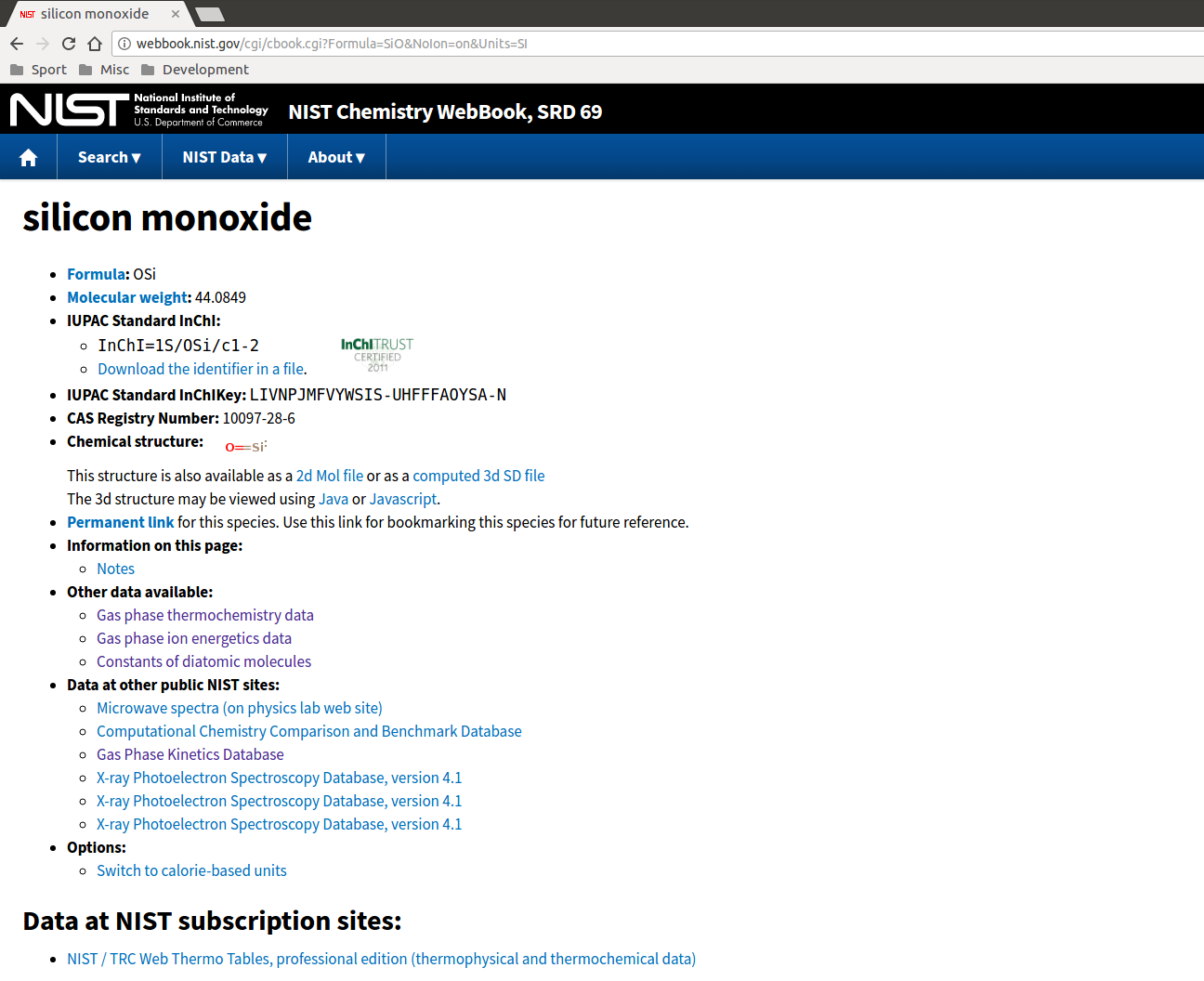
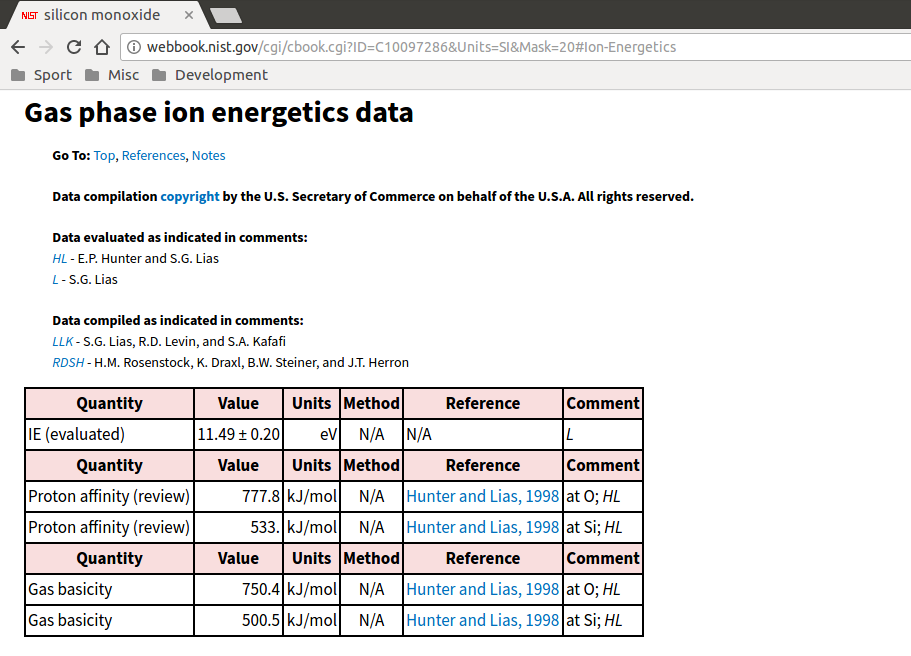
The first pieces of information needed are the ionisation energy (to turn SiO into SiO+) and the dissociation energy (to turn SiO into monatomic Si and O). The ionisation energy can be found on the “Gas phase ion energetics” link on the Chemistry WebBook species page and is usually in units of eV, so remember to convert:
The dissociation energy can be found either on the NIST Computational Chemistry Comparison and Benchmark Database (http://cccbdb.nist.gov/introx.asp), or alternatively from chemistry textbooks or other sources. In the case of SiO, the value is 798 kJ/mol.
We then need some information about the electronic ground state of the molecule, and its vibrational and rotational parameters. To get this, go back to the main page for SiO in the Chemistry WebBook and click on “Constants of diatomic molecules”. This gives the parameters of various energetic states of the molecule - we want the ground state with energy \(T_e=0\), so scroll all the way down to the entry at the very bottom:
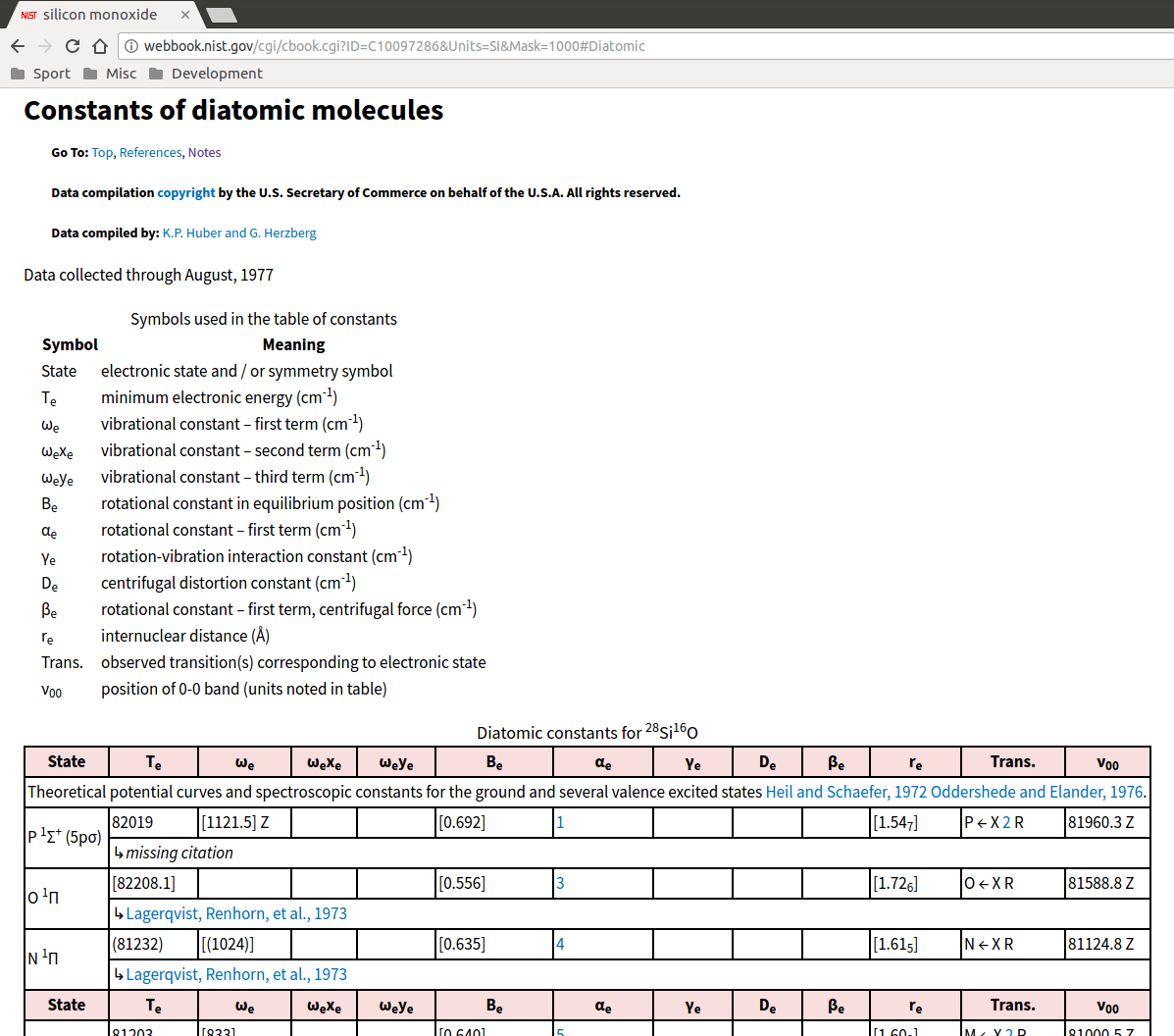
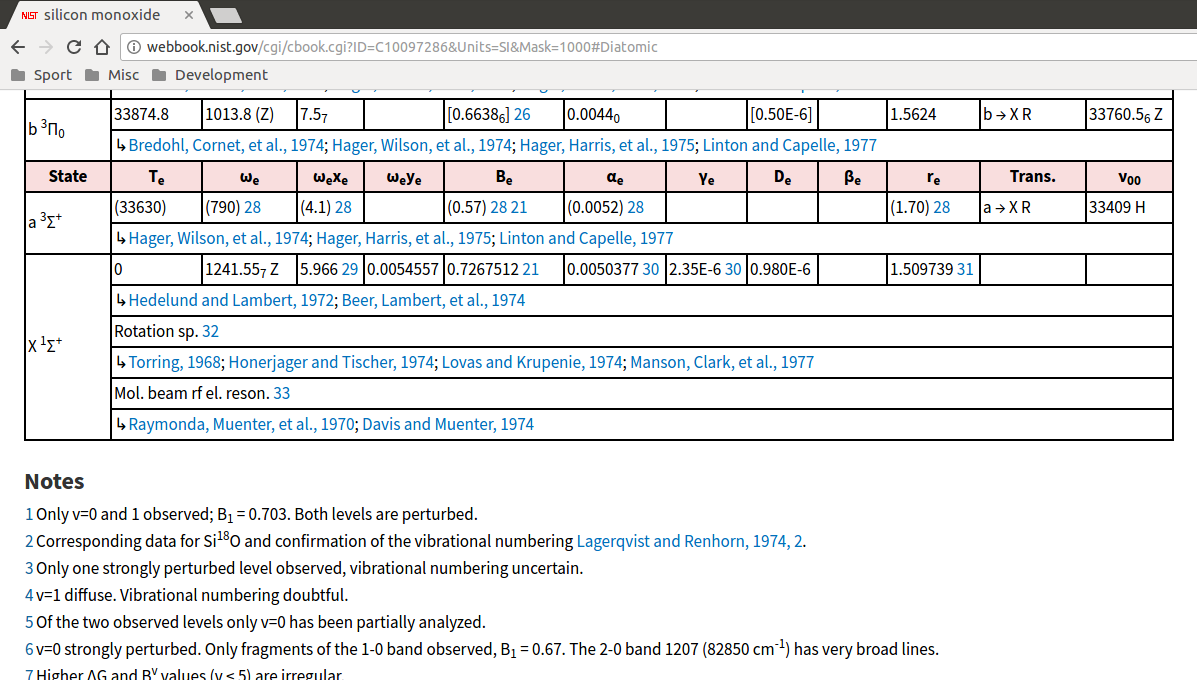
The multiplicity and electronic degeneracy of the ground state, \(g_0\), is generally given by the superscripted number in the molecular term expression in the “State” column - in the case of SiO, it’s 1.
Additional pieces of data needed to create a diatomic species object are:
The elemental stoichiometry of the species.
The molar mass of the species, in kg/mol.
The electric charge in units of the elementary charge.
The ionisation energy of the species, in J.
The dissociation (atomisation) energy of the species, in J, only used by neutral species.
The symmetry constant \(\sigma_s\), takes a value of 1 for heteronuclear molecules like SiO, and 2 for homonuclear molecules like \(O_2\).
The vibrational constant \(\omega_e\), in J.
The rotational constant \(B_e\), in J.
The polarisability of the species, in \(m^3\).
The multiplicity of the ground state.
The number of effective valence electrons, only used by neutral species (see docstring for more information).
Information about electron collision cross sections, only used by neutral species (see docstring for more information).
A list of radiation emission line data, if available (see Tutorial 1 for more information).
A list of sources describing the provenance of the data.
With these we have all the information to build a minplascalc data entry for the SiO species, which can be done by running the following code snippet:
from scipy import constants # type: ignore
import minplascalc as mpc
invcm_to_joule = constants.Boltzmann / (
0.01
* constants.physical_constants[
"Boltzmann constant in inverse meters per kelvin"
][0]
)
eV_to_joule = constants.elementary_charge
silicon_oxide = mpc.species.Diatomic(
name="SiO",
stoichiometry={"Si": 1, "O": 1},
molar_mass=0.044084905,
charge_number=0,
ionisation_energy=eV_to_joule * 11.49,
dissociation_energy=798 * 1000 / constants.Avogadro,
sigma_s=1,
g0=1,
w_e=invcm_to_joule * 1241.55,
b_e=invcm_to_joule * 0.7267512,
polarisability=3.962e-30,
multiplicity=1,
effective_electrons=10.495867769,
electron_cross_section=2.4197353806e-19,
emission_lines=[],
sources=[
"NIST Chemistry WebBook, NIST Standard Reference Database Number 69. "
"PJ Linstrom and WG Mallard (Editors), "
"National Institute of Standards and Technology, "
"Gaithersburg MD., http://webbook.nist.gov/chemistry/, "
"doi:10.18434/T4D303"
],
)
print(silicon_oxide)
silicon_oxide.to_file()
Species: SiO
Type: Diatomic molecule
Stoichiometry: {'Si': 1, 'O': 1}
Molar mass: 0.044084905 kg/mol
Charge number: 0
Dissociation energy: 1.3251101756047297e-18 J
Ionisation energy: 1.840900952466e-18 J
sigma_s: 1
g0: 1
w_e: 2.466272671803936e-20 J
B_e: 1.4436523891592904e-23 J
Polarisability: 3.962e-30 m^3
Multiplicity: 1
Effective valence electrons: 10.495867769
Electron cross section data: 2.4197353806e-19
Emission lines: 0
What’s happening here? First we import the minplascalc package, then we create a Diatomic species object using our data, and finally we save the contents to a file.
The Diatomic class constructor takes as arguments the list of data described above. Once created, the object can be written out to disk using the Species object’s to_file utility function, which saves data to a human-readable JSON-formatted file. It takes an optional file path argument - if omitted, the filename is created using the species’ name and written to the current working directory.
After this process it will be possible to create an SiO species object in any minplascalc calculation by importing it using either the explicit path to the JSON file, or (preferably) just the name of the species provided the JSON file is stored in any of the standard minplascalc data paths - see later demos for examples. Note that for this to work correctly in all cases, minplascalc’s JSON files must be stored on a case-sensitive operating system or partition.
Total running time of the script: (0 minutes 0.001 seconds)