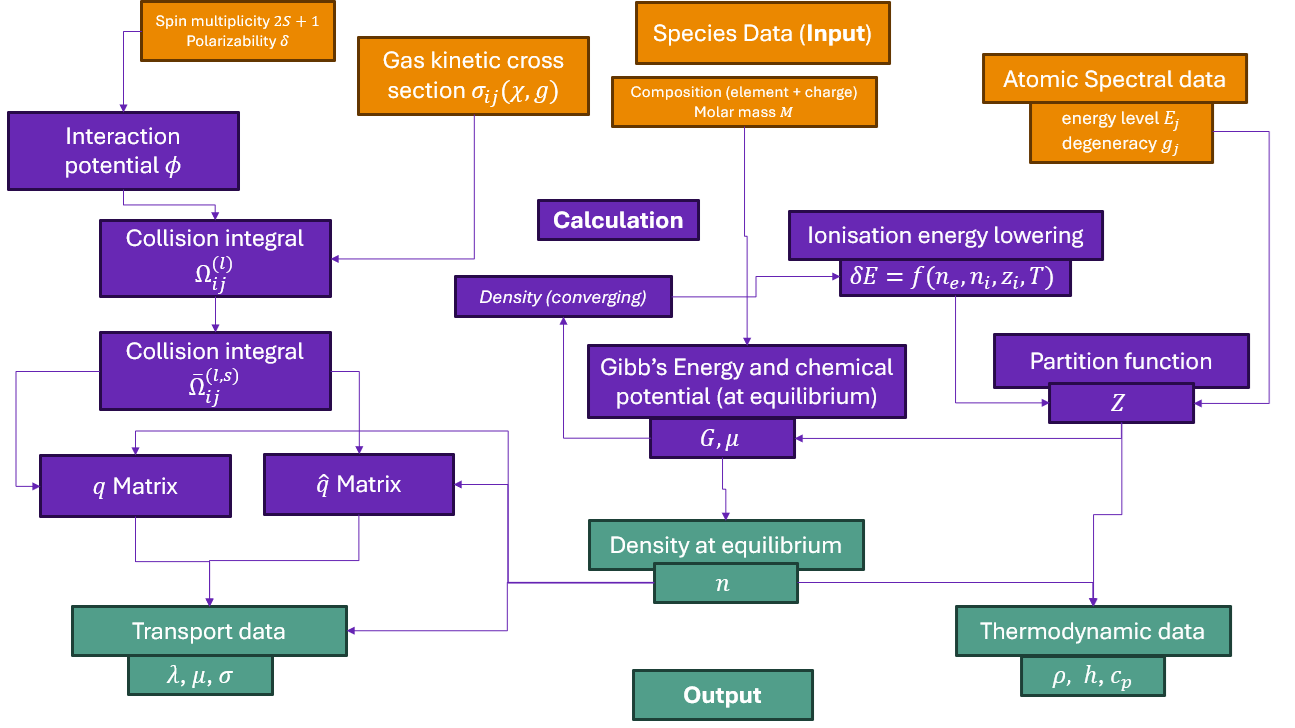
Tutorial 04: Calculating partition functions for individual species.#
Let’s use minplascalc to calculate and graph the internal and translational partition functions of various oxygen plasma species over a range of temperatures. To do this, just execute the following code snippets in order. The text in between describes what each piece of code is doing.
Import the required libraries.#
We start by importing the modules we need:
matplotlib for drawing graphs,
numpy for array functions,
and of course minplascalc.
import matplotlib.pyplot as plt
import numpy as np
import minplascalc as mpc
Create species objects for the species we’re interested in.#
Then, we create some species objects for different oxygen species using their names. This requires that the data files describing the species are stored in one of the minplascalc standard data storage paths.
species_names = ["O2", "O", "O+", "O++"]
species = [mpc.species.from_name(n) for n in species_names]
Define the range of temperatures we’re interested in.#
Next, we specify a range of temperatures we’re interested in calculating the partition functions at - in this case, from 1000 to 25000 K.
temperatures = np.linspace(1000, 25000, 100)
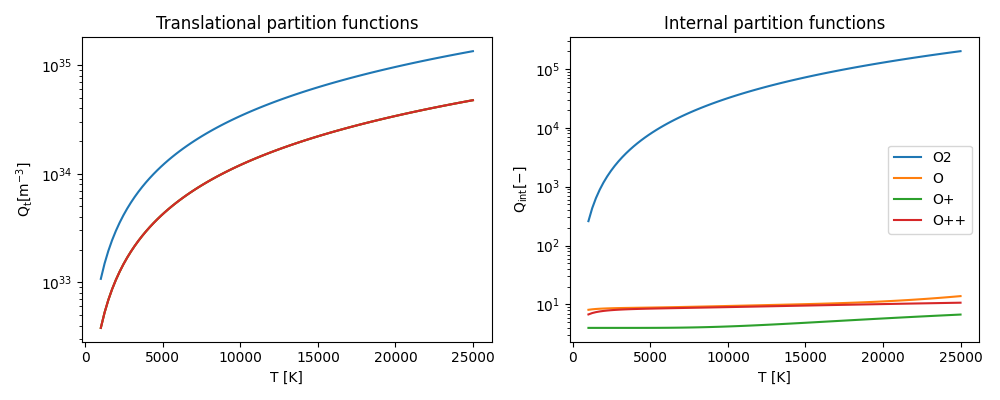
Calculate the partition functions for each species.#
Then calculate the actual partition functions, using the translational_partition_function (T) and internal_partition_function (T, \(\Delta E\)) functions of a minplascalc Species object.
The required arguments are T in K, and the ionisation energy lowering \(\Delta E\)) in J (here set to zero).
translational_partition_functions = [
[sp.translational_partition_function(T) for T in temperatures]
for sp in species
]
internal_partition_functions = [
[sp.internal_partition_function(T, 0) for T in temperatures]
for sp in species
]
Plot the results.#
Finally, to visualise the results, plot all the partition functions as a function of temperature over the range specified.
fig, axs = plt.subplots(1, 2, figsize=(10, 4))
ax = axs[0]
ax.set_title("Translational partition functions")
ax.set_xlabel("T [K]")
ax.set_ylabel(r"$\mathregular{Q_t [m^{-3}]}$")
for pf, sn in zip(translational_partition_functions, species_names):
ax.semilogy(temperatures, pf, label=sn)
ax = axs[1]
ax.set_title("Internal partition functions")
ax.set_xlabel("T [K]")
ax.set_ylabel(r"$\mathregular{Q_{int} [-]}$")
for pf, sn in zip(internal_partition_functions, species_names):
ax.semilogy(temperatures, pf, label=sn)
ax.legend()
plt.tight_layout()
Total running time of the script: (0 minutes 0.473 seconds)